HDX-NMR与HDX-MS在蛋白质结构动力学研究中的应用与进展
|
|
张媛媛, 汪鹏程, 李滔, 胡锐, 杨运煌, 刘买利
|
Development and Applications of HDX-NMR and HDX-MS in Protein Structure and Dynamics Research
|
|
ZHANG Yuanyuan, WANG Pengcheng, LI Tao, HU Rui, YANG Yunhuang, LIU Maili
|
|
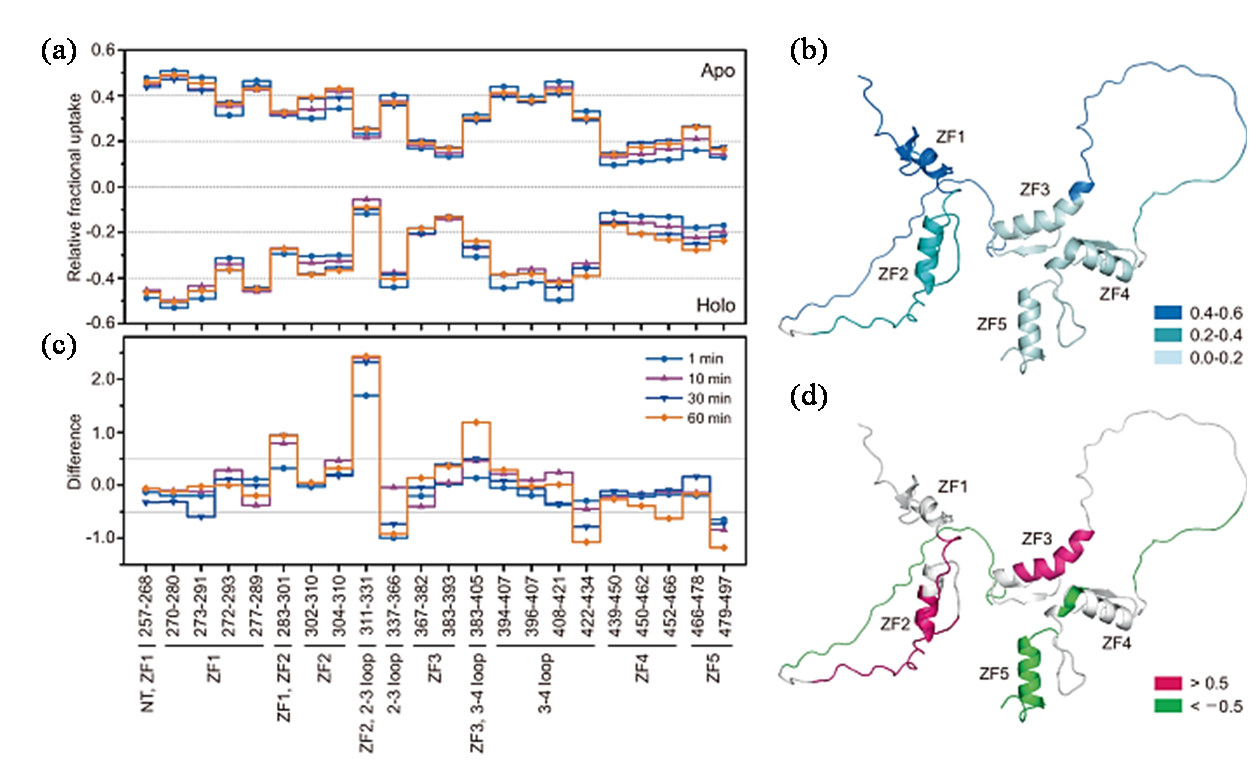
图4 HDX-MS揭示INSM1与核酸结合的高度动态构象特征[43]. (a) HDX-MS数据镜像图,对比了INSM1 ZF1-ZF5游离状态(Apo)与结合M2 DNA状态(Holo)所检测到的各肽段的相对氘摄取量;(b)将Apo状态的各肽段在1 min时的相对氘摄取量映射至INSM1 ZF1-ZF5结构上;(c) HDX-MS数据差异图,展示了Apo与Holo状态INSM1 ZF1-ZF5之间相对氘摄取量的变化;(d)氢氘交换60 min时,INSM1 ZF1-ZF5中Apo与Holo状态间存在显著差异的片段
|
Fig. 4 HDX-MS reveals the highly dynamic conformational characteristics of INSM1 in binding to nucleic acids[43]. (a) A mirror plot of HDX-MS data comparing the relative deuterium uptake for each peptide of free INSM1 ZF1-ZF5 (Apo) and INSM1 ZF1-ZF5 bound by M2 DNA (Holo); (b) The relative deuterium uptake for each peptide of Apo INSM1 ZF1-ZF5 at 1 min was mapped on the structure of INSM1 ZF1-ZF5; (c) A difference plot of HDX-MS data showing the changes in relative deuterium uptake between Apo and Holo INSM1 ZF1-ZF5; (d) The segments of INSM1 ZF1-ZF5 with significant changes between Apo and Holo states at 60 min
|
|
|
|
|

