
Chinese Journal of Magnetic Resonance ›› 2022, Vol. 39 ›› Issue (2): 184-195.doi: 10.11938/cjmr20212941
• Articles • Previous Articles Next Articles
Yan MA,Cang-ju XING,Liang XIAO*( )
)
Received:2021-08-19
Published:2022-06-05
Online:2021-11-16
Contact:
Liang XIAO
E-mail:xiaoliang@mail.buct.edu.cn
CLC Number:
Yan MA, Cang-ju XING, Liang XIAO. Knee Joint Image Segmentation and Model Construction Based on Cascaded Network[J]. Chinese Journal of Magnetic Resonance, 2022, 39(2): 184-195.
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
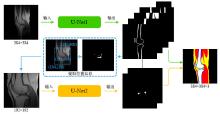
Fig.1
The overall block diagram of the proposed cascaded network. Let the coordinate of the lower left corner of the original image be (0, 0), (199, 204) is the center coordinate of the extracted cartilage and meniscus, (104, 109) and (295, 300) are the coordinates of the lower left and upper right corners of the crop area in the original image, respectively. The final result is obtained by merging the segmentation results of the two networks according to the extracted position information, and post-processing

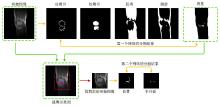
Fig.3
The first row is the initial image and the corresponding segmentation results of cortical bone, cancellous bone, muscle, fat, and background. Extract the cavity in the background and calculate its centroid to obtain the position information of cartilage and meniscus. Cut out with the centroid as the center to obtain a sub-image containing cartilage and meniscus, which is the input image of the second network. The second row respectively shows the cut-out schematic diagram and the segmentation results of cartilage and meniscus with the second network

Table 1
Analysis for the segmentation evaluation indicators of the proposed method and 4 comparison methods
| 评价指标 | 分割方法 | 统计参数 | 半月板 | 软骨 | 皮质骨 | 松质骨 | 脂肪 | 肌肉 |
| FPR | U-Net | Mean | 0.0003 | 0.0007 | 0.0054 | 0.0024 | 0.0091 | 0.0135 |
| Std | 0.0001 | 0.0002 | 0.0013 | 0.0007 | 0.0024 | 0.0033 | ||
| U-Net (focal loss) | Mean | 0.0002 | 0.0011 | 0.0093 | 0.0024 | 0.0074 | 0.0133 | |
| Std | 0.0001 | 0.0004 | 0.0019 | 0.0007 | 0.0019 | 0.0033 | ||
| U-Net (attention) | Mean | 0.0003 | 0.0010 | 0.0054 | 0.0027 | 0.0105 | 0.0122 | |
| Std | 0.0001 | 0.0003 | 0.0010 | 0.0007 | 0.0026 | 0.0035 | ||
| U-Net++ | Mean | 0.0004 | 0.0005 | 0.0054 | 0.0022 | 0.0089 | 0.0169 | |
| Std | 0.0001 | 0.0002 | 0.0010 | 0.0008 | 0.0022 | 0.0038 | ||
| 本文所提方法 | Mean | 0.0002 | 0.0013 | 0.0050 | 0.0023 | 0.0064 | 0.0132 | |
| Std | 0.0001 | 0.0004 | 0.0010 | 0.0008 | 0.0015 | 0.0035 | ||
| TPR | U-Net | Mean | 0.6347 | 0.5435 | 0.7550 | 0.9333 | 0.8799 | 0.9372 |
| Std | 0.1030 | 0.0912 | 0.0583 | 0.0297 | 0.0498 | 0.0264 | ||
| U-Net (focal loss) | Mean | 0.7291 | 0.6025 | 0.8053 | 0.9251 | 0.8547 | 0.9333 | |
| Std | 0.0824 | 0.1005 | 0.0466 | 0.0284 | 0.0638 | 0.0242 | ||
| U-Net (attention) | Mean | 0.7076 | 0.5773 | 0.7298 | 0.9375 | 0.8792 | 0.9231 | |
| Std | 0.0617 | 0.0794 | 0.0437 | 0.0336 | 0.0604 | 0.0244 | ||
| U-Net++ | Mean | 0.6883 | 0.5144 | 0.6800 | 0.9396 | 0.8459 | 0.9349 | |
| Std | 0.0926 | 0.0959 | 0.0494 | 0.0291 | 0.0600 | 0.0185 | ||
| 本文所提方法 | Mean | 0.7562 | 0.7291 | 0.7635 | 0.9410 | 0.8803 | 0.9422 | |
| Std | 0.0671 | 0.0793 | 0.0479 | 0.0250 | 0.0630 | 0.0231 | ||
| DCC | U-Net | Mean | 0.6755 | 0.5502 | 0.6980 | 0.9292 | 0.8868 | 0.9252 |
| Std | 0.0718 | 0.0499 | 0.0378 | 0.0272 | 0.0387 | 0.0224 | ||
| U-Net (focal loss) | Mean | 0.6884 | 0.5505 | 0.6608 | 0.9310 | 0.8860 | 0.9234 | |
| Std | 0.0520 | 0.0562 | 0.0396 | 0.0244 | 0.0491 | 0.0207 | ||
| U-Net (attention) | Mean | 0.6837 | 0.5551 | 0.6866 | 0.9289 | 0.8868 | 0.9232 | |
| Std | 0.0416 | 0.0416 | 0.0347 | 0.0273 | 0.0511 | 0.0235 | ||
| U-Net++ | Mean | 0.6389 | 0.5618 | 0.6502 | 0.9296 | 0.8730 | 0.9118 | |
| Std | 0.0568 | 0.0610 | 0.0333 | 0.0310 | 0.0529 | 0.0230 | ||
| 本文所提方法 | Mean | 0.7071 | 0.6023 | 0.7083 | 0.9351 | 0.8907 | 0.9312 | |
| Std | 0.0521 | 0.0396 | 0.0293 | 0.0234 | 0.0419 | 0.0176 | ||
| JAC | U-Net | Mean | 0.5394 | 0.4092 | 0.5391 | 0.8824 | 0.8102 | 0.8677 |
| Std | 0.0699 | 0.0491 | 0.0405 | 0.0319 | 0.0596 | 0.0289 | ||
| U-Net (focal loss) | Mean | 0.5484 | 0.4107 | 0.5069 | 0.8844 | 0.8055 | 0.8632 | |
| Std | 0.0528 | 0.0462 | 0.0421 | 0.0362 | 0.0698 | 0.0280 | ||
| U-Net (attention) | Mean | 0.5418 | 0.4077 | 0.5341 | 0.8772 | 0.8073 | 0.8653 | |
| Std | 0.0414 | 0.0379 | 0.0367 | 0.0337 | 0.0691 | 0.3033 | ||
| U-Net++ | Mean | 0.4931 | 0.3296 | 0.4954 | 0.8716 | 0.7851 | 0.8468 | |
| Std | 0.0511 | 0.0471 | 0.0415 | 0.0364 | 0.0699 | 0.0314 | ||
| 本文所提方法 | Mean | 0.5610 | 0.4572 | 0.5600 | 0.8896 | 0.8139 | 0.8761 | |
| Std | 0.0525 | 0.0373 | 0.0331 | 0.0300 | 0.0600 | 0.0251 | ||
| ASD/mm | U-Net | Mean | 1.3462 | 2.8589 | 1.2567 | 1.1638 | 1.3399 | 2.2038 |
| Std | 0.7835 | 0.8687 | 0.5117 | 0.4399 | 0.1666 | 0.6693 | ||
| U-Net (focal loss) | Mean | 1.3125 | 3.1638 | 1.2982 | 1.1325 | 1.3147 | 2.2202 | |
| Std | 0.6548 | 1.1781 | 0.2242 | 0.3914 | 0.3295 | 0.7109 | ||
| U-Net (attention) | Mean | 1.2539 | 2.7799 | 1.2440 | 1.5075 | 1.3017 | 2.4292 | |
| Std | 0.6181 | 0.8609 | 0.4551 | 0.3993 | 0.2916 | 1.3714 | ||
| U-Net++ | Mean | 1.6536 | 3.6479 | 1.6531 | 1.7551 | 1.4824 | 2.9900 | |
| Std | 0.6723 | 1.5050 | 1.5734 | 1.0557 | 0.2625 | 1.3447 | ||
| 本文所提方法 | Mean | 1.2470 | 2.6555 | 1.0716 | 1.0651 | 1.2412 | 1.9097 | |
| Std | 0.5749 | 0.8258 | 0.3542 | 0.3869 | 0.2607 | 0.5289 |
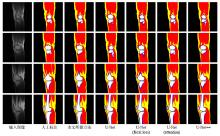
Fig.4
The comparison between the final segmentation results obtained by using the 5 automatic segmentation methods and the manual segmentation. From top to bottom, 4 consecutive equally spaced slices are collected for a volunteer image. Among them, fat, muscle, cancellous bone, cortical bone, cartilage, and meniscus are marked as yellow, red, white, blue, green, and pink, respectively
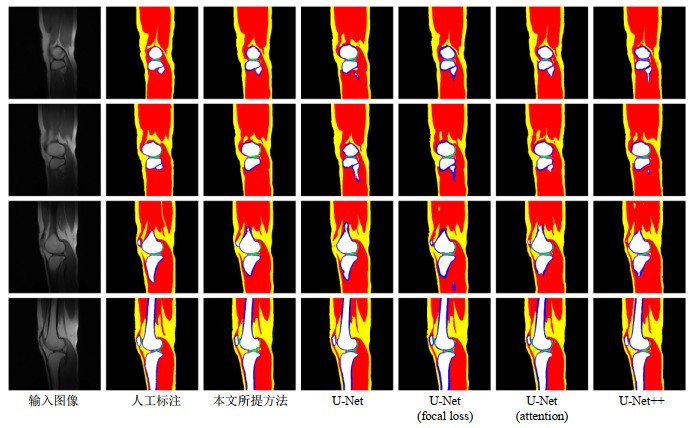
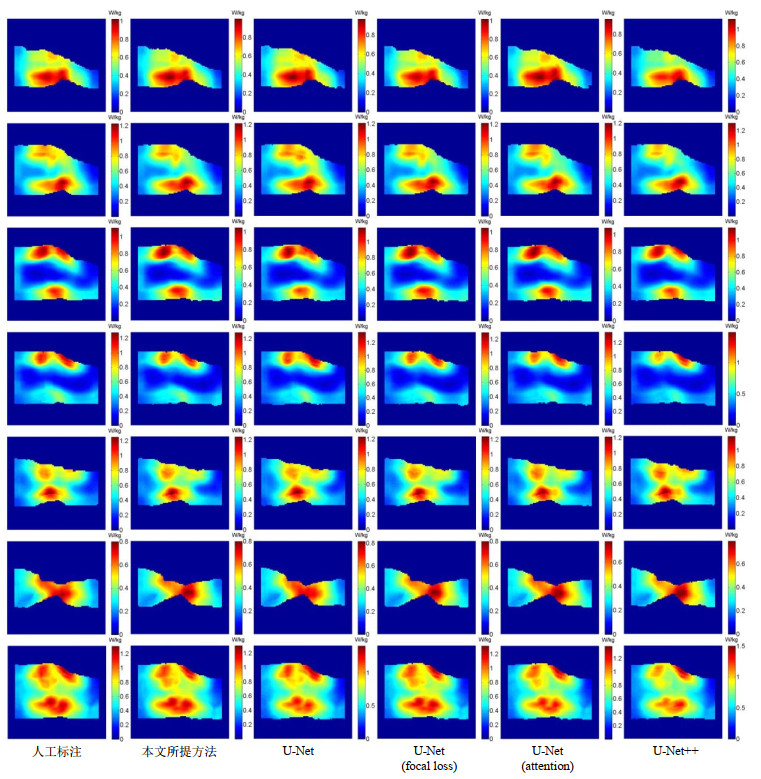
Fig.5
SAR10g distribution maps of a volunteer's knee joint. From left to right, they are based on the manual segmentation, and the models obtained by the proposed method, the U-Net using cross-entropy loss, the U-Net using focus loss, the U-Net using attention mechanism and U-Net++, respectively. The SAR10g values of the six models are 1.309 8 W/kg, 1.304 5 W/kg, 1.402 4 W/kg, 1.316 2 W/kg, 1.383 5 W/kg, 1.506 0 W/kg. The first row to the sixth row are the SAR10g distribution maps of 6 uniformly distributed slices, and the last row is the maximum density projection of the SAR10g
| 1 |
PADORMO F , BEQIRI A , HAJNAL J V , et al. Parallel transmission for ultrahigh-field imaging[J]. NMR Biomed, 2016, 29 (9): 1145- 1161.
doi: 10.1002/nbm.3313 |
| 2 |
GRAESSLIN I , HOMANN H , BIEDERER , et al. A specific absorption rate prediction concept for parallel transmission MR[J]. Magn Reson Med, 2012, 68 (5): 1664- 1674.
doi: 10.1002/mrm.24138 |
| 3 |
GRAESSLIN I , VERNICKEI P , BÖRNERT P , et al. Comprehensive RF safety concept for parallel transmission MR[J]. Magn Reson Med, 2015, 74 (2): 589- 598.
doi: 10.1002/mrm.25425 |
| 4 |
VAN DEN BERGEN B , VAN DEN BERG C A T , BARTELS L W , et al. 7 Tesla body MRI: B1 shimming with simultaneous SAR reduction[J]. Phys Med Biol, 2007, 52 (17): 5429- 5441.
doi: 10.1088/0031-9155/52/17/022 |
| 5 | ÖZTURKC N, ALBAYRAKS. Automatic segmentation of cartilage in high-field magnetic resonance images of the knee joint with an improved voxel-classification-driven region-growing algorithm using vicinity-correlated subsampling[J]. Comput Biol Med, 2016, 72, 90- 107. |
| 6 |
TANG J S , MILLINGTON S , ACTON S T , et al. Surface extraction and thickness measurement of the articular cartilage from MR images using directional gradient vector flow snakes[J]. IEEE Trans Biomed Eng, 2006, 53 (5): 896- 907.
doi: 10.1109/TBME.2006.872816 |
| 7 |
WILLIAMS T G , HOLMES A P , WATERTON J C , et al. Anatomically corresponded regional analysis of cartilage in asymptomatic and osteoarthritic knees by statistical shape modelling of the bone[J]. IEEE Trans Med Imaging, 2010, 29 (8): 1541- 1559.
doi: 10.1109/TMI.2010.2047653 |
| 8 |
SHAN L , ZACH C , CHARLES C , et al. Automatic atlas-based three-label cartilage segmentation from MR knee images[J]. Med Image Anal, 2014, 18 (7): 1233- 1246.
doi: 10.1016/j.media.2014.05.008 |
| 9 |
ZHANG K L , LU W M , MARZILIANO P . Automatic knee cartilage segmentation from multi-contrast MR images using support vector machine classification with spatial dependencies[J]. Magn Reson Imaging, 2013, 31 (10): 1731- 1743.
doi: 10.1016/j.mri.2013.06.005 |
| 10 | LONG J, SHELHAMER E, DARRELL T, et al. Fully convolutional networks for semantic segmentation[C]//IEEE Conference on Computer Vision and Pattern Recognition, 2015: 3431-3440. |
| 11 |
CHEN J C , PAPANDREOU G , KOKKIONS I , et al. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs[J]. IEEE T Pattern Anal, 2018, 40 (4): 834- 848.
doi: 10.1109/TPAMI.2017.2699184 |
| 12 |
AMBELLAN F , TACK A , EHLKE M , et al. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the osteoarthritis initiative[J]. Med Image Anal, 2019, 52, 109- 118.
doi: 10.1016/j.media.2018.11.009 |
| 13 |
LIU F , ZHOU Z Y , JANG H , et al. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging[J]. Magn Reson Med, 2018, 79 (4): 2379- 2391.
doi: 10.1002/mrm.26841 |
| 14 |
ZHOU Z Y , ZHAO G Y , KIJOWSKI R , et al. Deep convolutional neural network for segmentation of knee joint anatomy[J]. Magn Reson Med, 2018, 80 (6): 2759- 2770.
doi: 10.1002/mrm.27229 |
| 15 | RONNEBERGER O, FISCHER P, BROX T. U-Net: Convolutional networks for biomedical image segmentation[C]//International Conference on MICCAI 2015, 2015: 234-241. |
| 16 | DONG H, YANG G, LIU F D, et al. Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks[C]//MIUA 2017: Medical Image Understanding and Analysis, 2017: 506-517. |
| 17 | XIAO X, LIAN S, LUO Z M, et al. Weighted Res-UNet for high-quality retina vessel segmentation[C]//9th International Conference on Information Technology in Medicine and Education (ITME), Hang Zhou, 2018: 327-331. |
| 18 |
WENG Y , ZHOU T B , LI Y J , et al. NAS-Unet neural architecture search for medical image segmentation[J]. IEEE Access, 2019, 7, 44247- 44257.
doi: 10.1109/ACCESS.2019.2908991 |
| 19 | NORMAN B , PEDOIA V , MAJUMDAR S , et al. Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry[J]. Radiology, 2018, 288 (2): 177- 185. |
| 20 |
AMBELLAN F , TACK A , EHLKE M , et al. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks[J]. Med Image Anal, 2019, 52, 109- 118.
doi: 10.1016/j.media.2018.11.009 |
| 21 |
BYRA M , WU M , ZHANG X D , et al. Knee menisci segmentation and relaxometry of 3D ultrashort echo time cones MR imaging using attention U-Net with transfer learning[J]. Magn Reson Med, 2020, 83 (3): 1109- 1122.
doi: 10.1002/mrm.27969 |
| 22 | XIAO L , LOU Y K , ZHOU H Y . A U-Net network-based rapid construction of knee models for specific absorption rate estimation[J]. Chinese J Magn Reson, 2020, 27 (2): 144- 151. |
| 肖亮, 娄煜堃, 周航宇. 用于SAR估计的基于U-Net网络的快速膝关节模型重建[J]. 波谱学杂志, 2020, 27 (2): 144- 151. | |
| 23 |
LIN T Y , GOYAL P , GIRSHICK R , et al. Focal loss for dense object detection[J]. IEEE T Pattern Anal, 2020, 42 (2): 318- 327.
doi: 10.1109/TPAMI.2018.2858826 |
| 24 |
HOMANN H , BÖRNERT P , EGGERS H , et al. Toward individualized SAR models and in vivo validation[J]. Magn Reson Med, 2011, 66 (6): 1767- 1776.
doi: 10.1002/mrm.22948 |
| 25 |
WOLF S , DIEHL D , GEBHARDT M , et al. SAR simulations for high-field MRI: how much detail, effort, and accuracy is needed?[J]. Magn Reson Med, 2013, 69 (4): 1157- 1168.
doi: 10.1002/mrm.24329 |
| 26 |
COLLINS C M , SMITH M B . Spatial resolution of numerical models of manand calculated specific absorption rate using the FDTD method: a study at 64 MHz in a magnetic resonance imaging coil[J]. Magn Reson Imaging, 2003, 18 (3): 383- 388.
doi: 10.1002/jmri.10359 |
| 27 |
PETERSON D M , CARRUTHERS C E , WOLVERTON B L , et al. Application of a birdcage coil at 3 Tesla to imaging of the human knee using MRI[J]. Magn Reson Med, 1999, 42 (2): 215- 221.
doi: 10.1002/(SICI)1522-2594(199908)42:2<215::AID-MRM1>3.0.CO;2-8 |
| 28 | BYRA M , WU M , ZHANG X D , et al. Knee menisci segmentation and relaxometry of 3D ultrashort echo time cones MR imaging using attention U‐Net with transfer learning[J]. Magn Reson Med, 2019, 83 (3): 1109- 1122. |
| 29 | ZHOU Z W, SIDDIQUEE M M R, TAJBAKHSH N, et al. Unet++: A nest u-net architecture for medical image segmentation[C]//DLMIA 2018, ML-CDS 2018: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, 2018, 11045: 3-11. |
| 30 | LIU K W , LIU Z L , WANG X Y , et al. Prostate cancer diagnosis based on cascaded convolutional neural networks[J]. Chinese J Magn Reson, 2020, 37 (2): 152- 161. |
| 刘可文, 刘紫龙, 汪香玉, 等. 基于级联卷积神经网络的前列腺磁共振图像分类[J]. 波谱学杂志, 2020, 37 (2): 152- 161. |
| [1] | MA Yingxue, ZHAO Yanqiang, YANG Xiaodong, JIANG Bin, TAO Cheng. Opportunities and Challenges of High-field and Ultra-high-field Magnetic Resonance Imaging in China [J]. Chinese Journal of Magnetic Resonance, 2025, 42(3): 334-344. |
| [2] | SUI Meiju, ZHANG Lei, WANG Ruifang, LUO Yingying, LI Sha, QIU Maosong, XU Qiuyi, CHEN Daiqin, CHEN Shizhen, ZHOU Xin. MRI-traceable Nanoenzyme for Cascade Catalysis-enhanced Immunotherapy [J]. Chinese Journal of Magnetic Resonance, 2025, 42(3): 231-248. |
| [3] | CHEN Qun, YANG Zijian, CHENG Xinyi, JIA Siyi, DU Xiaoxia, WANG Mengxing. Application of Magnetic Resonance Imaging Technology in Pediatric Exercise Intervention Research [J]. Chinese Journal of Magnetic Resonance, 2025, 42(2): 195-204. |
| [4] | PANG Qifan, WANG Zhichao, WU Yupeng, LI Jianqi. The Impact of K-Space Filling Strategy on Fat Artifacts in APT Imaging Based on FLASH Sequence [J]. Chinese Journal of Magnetic Resonance, 2024, 41(4): 443-453. |
| [5] | XU Zhenshun, YUAN Xiaohan, HUANG Ziheng, SHAO Chengwei, WU Jie, BIAN Yun. Multi-source Feature Classification Model of Pancreatic Mucinous and Serous Cystic Neoplasms Based on Deep Learning [J]. Chinese Journal of Magnetic Resonance, 2024, 41(1): 19-29. |
| [6] | LIU Ying, LIN Ling, YUAN Binhua, ZHANG Haowei. Research Progress of MRI Gradient Waveform Generator [J]. Chinese Journal of Magnetic Resonance, 2024, 41(1): 99-115. |
| [7] | REN Hongjin, MA Yan, XIAO Liang. Knee Joint Model Construction and Local Specific Absorption Rate Estimation Based on Generative Adversarial Networks [J]. Chinese Journal of Magnetic Resonance, 2023, 40(4): 410-422. |
| [8] | LI Pan,FANG Delei,ZHANG Junxia,MA Debei. Magnetic Resonance Compatibility Analysis Method of Surgical Robotic System Based on Image Quality Evaluation [J]. Chinese Journal of Magnetic Resonance, 2023, 40(1): 79-91. |
| [9] |
De-gang TANG,Hong-chuang LI,Xiao-ling LIU,Lei SHI,Hai-dong LI,Chao-hui YE,Xin ZHOU.
A Simulation Study on the Effect of the High Permittivity Materials Geometrical Structure on the Transmit Field |
| [10] | Zhen-yu WANG, Ying-shan WANG, Jin-ling MAO, Wei-wei MA, Qing LU, Jie SHI, Hong-zhi WANG. Magnetic Resonance Images Segmentation of Synovium Based on Dense-UNet++ [J]. Chinese Journal of Magnetic Resonance, 2022, 39(2): 208-219. |
| [11] | Jun LUO, Sheng-ping LIU, Xing YANG, Jia-sheng WANG, Ye LI. Design of a 5 T Non-magnetic Magnetic Resonance Radio Frequency Power Amplifier [J]. Chinese Journal of Magnetic Resonance, 2022, 39(2): 163-173. |
| [12] | Ju-min ZHANG,Shi-zhen CHEN,Xin ZHOU. Dual-modal MRI T1-T2 Contrast Agent Based on Dynamic Organic Gadolinium Nanoparticles [J]. Chinese Journal of Magnetic Resonance, 2022, 39(1): 11-19. |
| [13] | Zhi-chao WANG,Ji-lei ZHANG,Yu ZHAO,Ting HUA,Guang-yu TANG,Jian-qi LI. CEST Imaging of the Abdomen with Neural Network Fitting [J]. Chinese Journal of Magnetic Resonance, 2022, 39(1): 33-42. |
| [14] | Han-wei WANG,Hao WU,Jing TIAN,Jun-feng ZHANG,Peng ZHONG,Li-zhao CHEN,Shu-nan WANG. The Diagnostic Value of Quantitative Parameters of T2/FLAIR Mismatch Sign in Evaluating the Molecular Typing of Lower-grade Gliomas [J]. Chinese Journal of Magnetic Resonance, 2022, 39(1): 56-63. |
| [15] | Long XIAO,Xiao-lei ZHU,Ye-qing HAN,Shi-zhen CHEN,Xin ZHOU. Design and Application of Micellar Magnetic Resonance Imaging Molecular Probe [J]. Chinese Journal of Magnetic Resonance, 2021, 38(4): 474-490. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||